
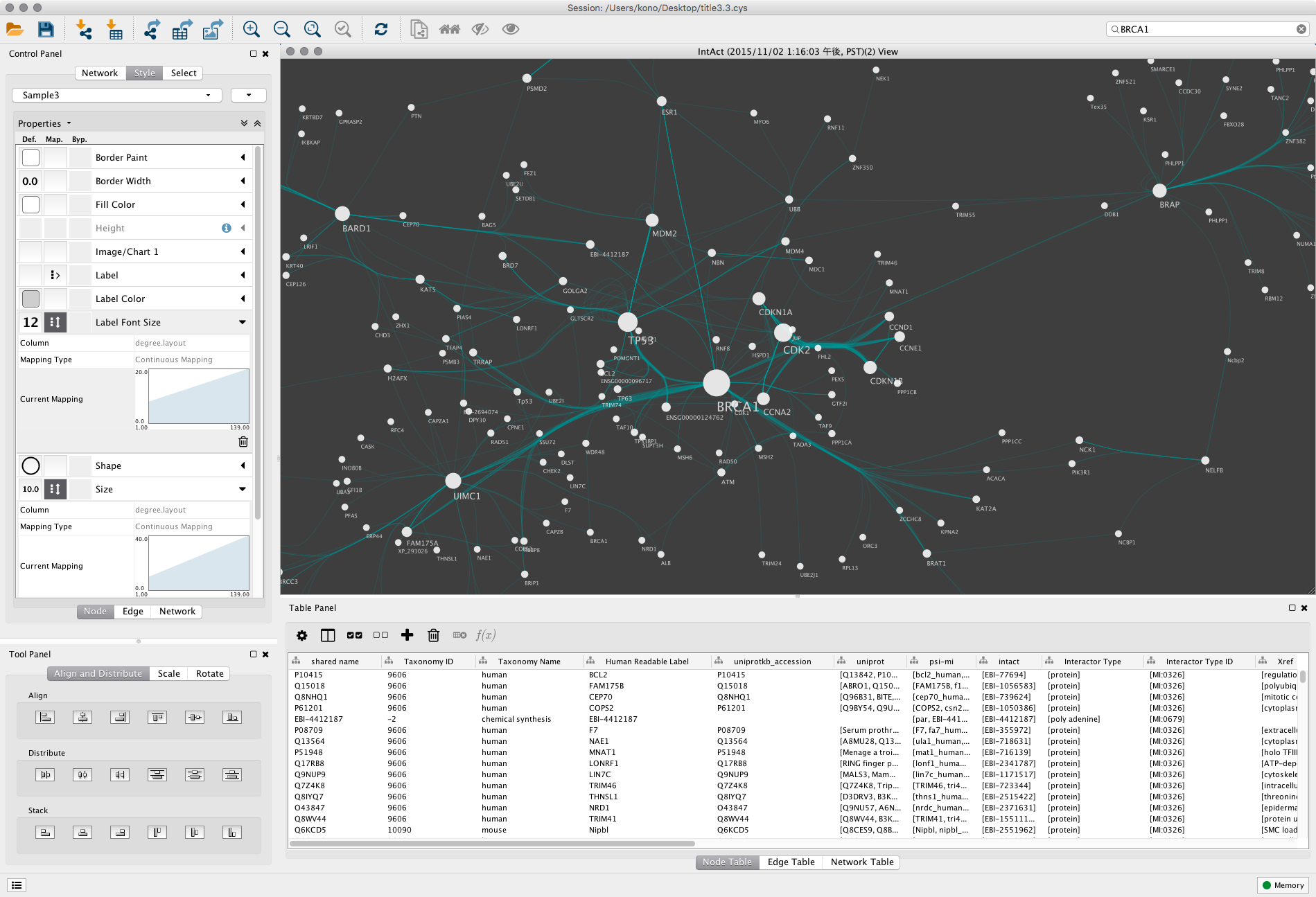
Metabolic perturbations can be investigated on the levels of genomic topology, gene expression or proteomics, aided by functional ontology interpretation or pathway mapping. However, the exact biochemical changes in different organs of ETS-exposed animal models are not understood on the systems level.Īmong the functional genomics technologies, metabolomics may better assist in understanding physiology on systems level, because the metabolome is the ultimate outcome of biochemical networks and close to disease phenotypes. Environmental tobacco smoke exposure has also been associated with abnormal fetal development. Biochemical studies suggest that ETS can alter cell signaling and metabolic functions that can impair normal cellular growth and morphology in lung tissues. Įxposure to environmental tobacco smoke (ETS) during fetal development can cause serious health consequences in later stages of life to increase the risk of respiratory disease and susceptibility. MetaMapp graphs efficiently visualizes mass spectrometry based metabolomics datasets as network graphs in Cytoscape, and highlights metabolic alterations that can be associated with higher rate of pulmonary diseases and infections in children prenatally exposed to ETS. Fetuses from ETS-exposed dams expressed lower lipid and nucleotide levels and higher amounts of energy metabolism intermediates than control animals, indicating lower biosynthetic rates of metabolites for cell division, structural proteins and lipids that are critical for in lung development.
Cytoscape visualization of differential statistics results using these graphs showed that overall, fetal lung metabolism was more impaired than lungs and blood metabolism in dams. MetaMapp graphs in Cytoscape showed much clearer metabolic modularity and complete content visualization compared to conventional biochemical mapping approaches. In fetal and maternal lungs, and in maternal blood plasma from pregnant rats exposed to environmental tobacco smoke (ETS), 459 unique metabolites comprising 179 structurally identified compounds were detected by gas chromatography time of flight mass spectrometry (GC-TOF MS) and BinBase data processing. We present a novel approach to integrate biochemical pathway and chemical relationships to map all detected metabolites in network graphs (MetaMapp) using KEGG reactant pair database, Tanimoto chemical and NIST mass spectral similarity scores. Hence, network visualizations that rely on current biochemical databases are incomplete and also fail to visualize novel, structurally unidentified metabolites. However, mass spectrometry-based non-targeted metabolomics datasets often comprise many metabolites for which links to enzymatic reactions have not yet been reported. Genome- reconstructed metabolic pathways may be used to map and interpret dysregulation in metabolic networks. To study the biochemical changes that may precede lung diseases, metabolomic effects on fetal and maternal lungs and plasma from rats exposed to ETS were compared to filtered air control animals. Let us know how this access is important for you.Exposure to environmental tobacco smoke (ETS) leads to higher rates of pulmonary diseases and infections in children. Many UC-authored scholarly publications are freely available on this site because of the UC's open access policies.
#Cytoscape visualization code
Availability and implementationImplementation available at, while the source code is available at. Finally, the application supports automation of its main features via Cytoscape commands. Users can also filter networks and click on nodes and edges to access all their related details. The app offers three visualizations: one only displaying interactions, another representing every evidence and the last one emphasizing evidence where mutated versions of proteins were used. Users can query a network by providing its molecules, identified by different fields and optionally include all their interacting partners in the resulting network.
It build networks where nodes are interacting molecules (mainly proteins, but also genes, RNA, chemicals…) and edges represent evidence of interaction.

SummaryIntAct App is a Cytoscape 3 application that grants in-depth access to IntAct's molecular interaction data.


 0 kommentar(er)
0 kommentar(er)
